PROMISE – PROduction of tailored MIcrobial oil using Sustainable platforms and process Engineering
Extensive animal agriculture as it is now performed in many regions of the world is generally harmful to the planet for several reasons, including environmental aspects, health considerations and ethical concerns. Alternatives such as plant-based products are increasing in relevance but have some disadvantages compared to animal products. The main reason is the difference in texture, bioavailability of nutrients and the difference in taste. Fat plays an important role in enhancing flavor, texture and mouthfeel, contributing to creaminess and satisfaction in foods like baked goods, sauces and spreads. By adjusting fat profiles, the new food products can closely mimic the taste of the original animal product. Furthermore, considering that fat is a significant contributor to health concerns such as cardiovascular diseases, it is important to be able to modulate the fat content. To achieve this, an efficient process for producing tailored fat profiles is required. Besides producing fat tissue by cell culture/tissue engineering, microbial processes may be used with the well-known benefits such as fast and efficient growth, potential for high yields and easy genetic accessibility. By exploiting the advantages of microbial systems for lipid production, a tailored fatty acid composition can be produced to ensure certain taste profiles while keeping health benefits in mind.
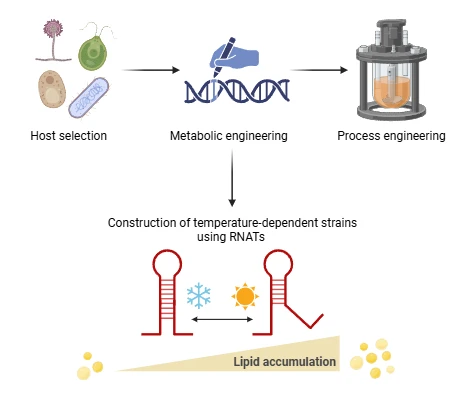
This research aims to develop a precision fermentation platform for the efficient and customizable production of microbial lipids. By strategically integrating temperature-responsive genetic elements into fat metabolism and combining them with process control strategies based on temperature variations and feeding regimes, the production process can be guided along a desired trajectory. The project also includes application testing for food, exploring the use of these microbial lipids in products such as mayonnaise and cultured meat.
If you are an ambitious, passionate, and highly motivated student interested in expanding your knowledge in this field, please contact atena.galefi(at)tum.de for further details. Applications for Bachelor's and Master's theses and research internships are welcome.
PRIMO: Precision Regulation of Interfaced Microbial Quorum Sensing for Optimized Food Protein Production
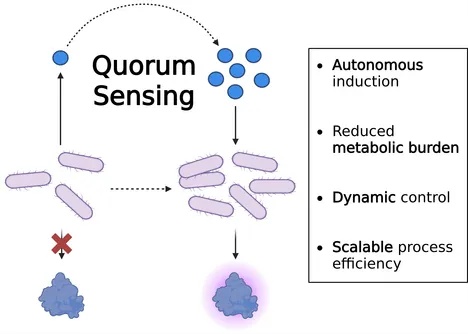
Harnessing bacterial quorum sensing (QS) opens new possibilities for molecular process control and precision fermentation. By allowing microbes to regulate gene expression in response to their own population density, QS enables dynamic, self-regulating production processes without the need for external inducers.
Our project focuses on the endogenous ComQXPA system of Bacillus subtilis. This microorganism is widely used in industrial fermentation, and in many applications classified as generally recognized as safe (GRAS), with several of its enzymes and metabolites approved for food and biotechnological applications.
We explore how this natural communication system can drive the efficient production of functional food proteins. Key challenges include ensuring robust signal transduction and achieving scalability without loss of performance.
Self-regulating QS-based systems have the potential to reduce metabolic burden, enhance process efficiency, and scale sustainable protein production. By utilizing autonomous control, this approach could establish a new tool for heterologous protein production.
If you are an ambitious, passionate, and highly motivated student interested in expanding your knowledge in this area of research, please contact nicholas.boehnlein(at)tum.de for further details. Applications for Bachelor's and Master's theses and research internships are welcome.
EggSPLORE: Egg Structural and function Protein Library using Optimized Recombinant Expression
The increasing demand for high-quality, animal-free proteins and concerns about the environmental impact of conventional egg production methods have led to the need for research into recombinant egg proteins. To date, plant-based replacements have not been able to fully replicate the functional properties of egg proteins, which play a crucial role in providing nutrients, antimicrobial and antioxidant properties, as well as physicochemical properties and texture in food products. Developing efficient methods for scalable production of recombinant egg proteins offer the potential to meet this demand and contribute to our future bioeconomy.
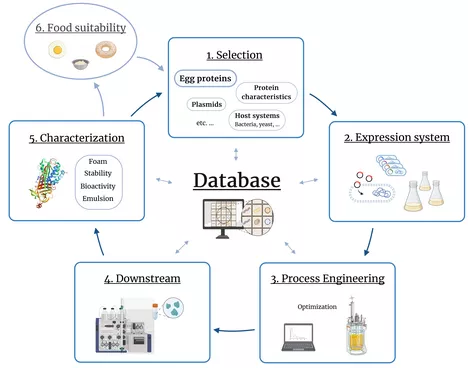
The EggSPLORE project aims to establish a platform for the recombinant production of selected egg yolk and egg white proteins in microbial systems. Explicitly, bacterial systems will be investigated due to their high potential in terms of efficiency and space-time yield for this group of proteins.
Core elements of this project include:
Development of successful expression systems with suitable production hosts and plasmids
Scale-up to a fully characterised bioreactor production process and its optimisation using model-based methods and adaptation of process control
Purification of the produced proteins and characterisation for their functional properties and suitability for use in food formulations
If you are an ambitious, passionate, and highly motivated student interested in working on cloning and bacterial cultivation or mathematical modeling of biological pathways, please contact fr.beck(at)tum.de for further details. Applications for Bachelor's and Master's theses and research internships are welcome.
FORTUNE: Fermentation Optimization using RNA-Thermometers for Upgraded Nutrient Efficiency
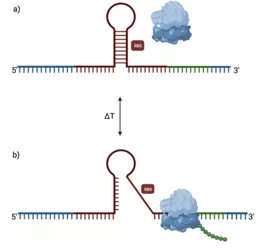
a) Active regulatory mechanism that masks RBS, b) Temperature-induced conformational change favoring translation. Created with BioRender.com
The global food supply chain is facing several challenges due to population growth and climate change and its consequences. As a result, increased interest in alternative food production has led to a re-evaluation of established production processes based on fermentation. A new approach, called precision fermentation, is revolutionizing the food sector by diversifying the range of applications and products by developing optimized metabolic pathways and assembling the genes involved in microbial chassis. To achieve this, genetically modified organisms (GMOs) are being optimized for the heterologous production of alternative food proteins. A key avenue for improving the efficiency of bioprocesses is to decouple biomass and product formation, addressing the challenge posed by the inefficiency of recycling biomass, a common by-product.
For this task, RNA thermometers (RNAT) can be used as gene expression 'dimmer switches' for novel fermentation products. Typically located within the 5' untranslated region (5'UTR), RNAT adopts a hairpin-like structure that effectively masks the ribosome binding site (RBS) (Figure 1a). In response to temperature changes, conformational changes are induced leading to exposure of the RBS and initiation of translation (Figure 1b).

Therefore, the evaluation of an RNAT-based expression system at different laboratory scales and the establishment of a reference system with in-depth characterization using common bioprocessing parameters and methods is key (Figure 2).
Combining precision fermentation with a highly temperature-controlled bacterial RNAT-based expression system could lead to an efficient and optimized production process for novel, specific applications, such as animal-free proteins.
If you are an ambitious, passionate, and highly motivated student interested in working on cloning and bacterial cultivation or mathematical modeling of biological pathways, please contact christina.peternell(at)tum.de for further details. Applications for Bachelor's and Master's theses and research internships are welcome.